Whole-Genome Sequencing (WGS)
What is whole-genome sequencing?
Whole-genome sequencing (WGS) is a comprehensive method for analyzing entire genomes. Genomic information has been instrumental in identifying inherited disorders, characterizing the mutations that drive cancer progression, and tracking disease outbreaks. Rapidly dropping sequencing costs and the ability to produce large volumes of data with today’s sequencers make whole-genome sequencing a powerful tool for genomics research.
While this method is commonly associated with sequencing human genomes, the scalable, flexible nature of next-generation sequencing (NGS) technology makes it equally useful for sequencing any species, such as agriculturally important livestock, plants, or disease-related microbes.

Advantages of whole-genome sequencing
- Provides a high-resolution, base-by-base view of the genome
- Captures both large and small variants that might be missed with targeted approaches
- Identifies potential causative variants for further follow-up studies of gene expression and regulation mechanisms
- Delivers large volumes of data in a short amount of time to support assembly of novel genomes
An uncompromised view of the genome
Unlike focused approaches such as exome sequencing or targeted resequencing, which analyze a limited portion of the genome, whole-genome sequencing delivers a comprehensive view of the entire genome. It is ideal for discovery applications, such as identifying causative variants and novel genome assembly.
Whole-genome sequencing can detect single nucleotide variants, insertions/deletions, copy number changes, and large structural variants. Due to recent technological innovations, the latest genome sequencers can perform whole-genome sequencing more efficiently than ever.
Compare whole-genome and exome sequencing
Explore the benefits of each approach to determine which method is best for your research.
Emerging applications and advances in whole-genome sequencing
Illumina Distinguished Scientist Gary Schroth discusses the latest advances in Illumina whole-genome sequencing solutions and the vast potential of emerging genomics technologies.
Access WebinarKey Whole-Genome Sequencing Methods
Large whole-genome sequencing
Sequencing large genomes (> 5 Mb), such as human, plant, or animal genomes, can provide valuable information for disease research and population genetics.
Small whole-genome sequencing
Small genome sequencing (≤ 5 Mb) involves sequencing the entire genome of a bacterium, virus, or other microbe. Without requiring bacterial culture, researchers can sequence thousands of small organisms in parallel using NGS.
De novo sequencing
De novo sequencing refers to sequencing a novel genome where there is no reference sequence available. NGS enables fast, accurate characterization of any species.
Phased sequencing
Phased sequencing, or genome phasing, distinguishes between alleles on homologous chromosomes, resulting in whole-genome haplotypes. This information is often important for genetic disease studies.
Human whole-genome sequencing
Previously a challenging application, human whole-genome sequencing has never been simpler. It offers the most detailed view into our genetic code.
Long-read sequencing
Long reads can help resolve challenging regions of the genome, such as those containing highly variable or highly repetitive elements.
Featured whole-genome sequencing workflow
This user-friendly three-step WGS workflow provides a fully featured, rapid solution for labs and delivers high-quality insights across the entire genome for all variant classes.
Library Preparation
Sequencing
Data Analysis
Featured Products

Multiomics multiplies your discovery power
See how you can use multiomics to better connect genotype to phenotype and obtain a full cellular readout not found through single omics approaches.
Learn About Multiomics at a Glance
How Scientists Use WGS

Investigating the genetics of COVID-19 susceptibility
Illumina is providing whole-genome sequencing for a UK-wide study led by Genomics England, designed to compare the genomes of severely and mildly ill COVID-19 patients.
The time is now for microbiome studies
Whole-genome shotgun sequencing and transcriptomics provide researchers and pharmaceutical companies with data to refine drug discovery and development.

Analyzing leukemia samples with WGS
A New England Journal of Medicine study found that using WGS to assess leukemia samples produced more accurate results, in less time, than karyotyping or fluorescence in situ hybridization.
Featured Webinars
Emerging applications and advances in WGS
Illumina Distinguished Scientist Gary Schroth discusses the latest WGS advances and the vast potential of emerging genomics technologies.
Advancements in whole-genome sequencing
Watch how improvements in informatics are advancing whole-genome sequencing, and find out the latest on Illumina Complete Long Reads.
From sample to genomic insight
See what Mile Lelivelt, VP of Product Managment for Software and Informatics, shares about Illumina solutions to arrive at critical genomic insights.
Additional Resources
Sequencing platforms
Compare sequencing platforms by application and specification. Find tools and guides to help you choose the right instrument.

Accurate CYP2D6 genotyping using WGS data
WGS offers a promising option for building accurate drug-metabolizing enzyme allele frequency databases for pharmacogenomics.
Library Prep and Array Kit Selector
Determine the best kit for your needs based on your project type, starting material, and method of interest.

Sequencing services
Find high-quality whole-genome and other sequencing services that deliver analyzed data to researchers.

Sequencing Method Explorer
Use this interactive tool to explore experimental NGS library preparation methods compiled from the scientific literature.
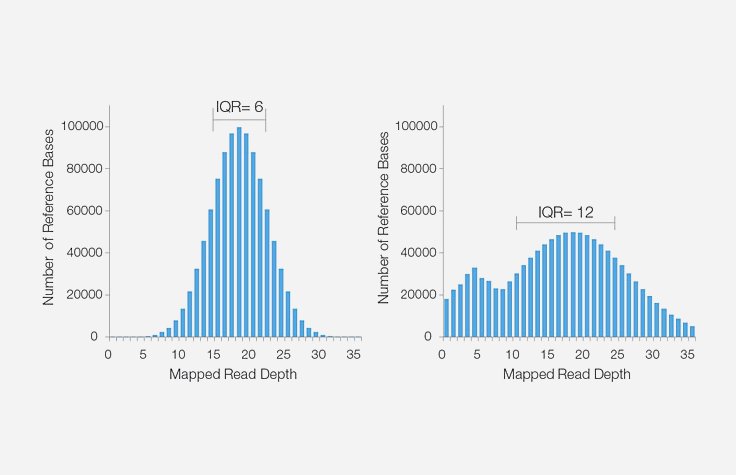
Sequencing coverage tips
Learn how to estimate and achieve the necessary sequencing coverage for your experiment.
Introducing Illumina Complete Long Reads
This novel technology will bright more light to even the darkest corners of the genome. Illumina Complete Long Reads helps resolve the most challenging regions of the genome and makes long-read sequencing accessible and streamlined by enabling short and long reads from a single platform.
Learn about the technology and products
Additional Tips and Training Opportunities
Find content and products for your field
The user-friendly "Recommended Links" feature allows you to quickly find in-depth content and products relevant to your specific field of interest. You can access this option from the top of any illumina.com page.
What is the PhiX Control v3 Library?
This library is derived from the small, well-characterized bacteriophage genome, PhiX. It is an ideal sequencing control for run quality monitoring.
Interested in receiving newsletters, case studies, and information on whole-genome sequencing?
Enter your email address.